Updated: January 19th, 2026
Overview
Researchers and policy makers can use the genetic evolution of viruses to make critical epidemiological decisions, aiding in understanding emerging disease trends and informing policy responses to outbreaks 3. Often, however, these analyses are conducted in isolation, with results and data rarely made public until after publication, and they do not always capture how viral evolution unfolds over time 1. In response to this bottleneck, collaborators from the Fred Hutchinson Cancer Research Center and the University of Basel launched Nextstrain in 2015 4.
This platform provides near real-time dashboards for viral diseases of high public health interest—from endemic infections (seasonal influenza, dengue) to emerging outbreaks (avian influenza, Zika, Ebola)—plus access to the underlying bioinformatics tools 3. Its framework enables ongoing monitoring of pathogen evolution and can expand to additional diseases developed and maintained by the research community. This approach allows robust, reproducible, rapid analysis and dissemination of results that keep pace with sequencing data generation 1,2.
Data for Nextstrain maintained dashboards are sourced from public repositories such as the National Center for Biotechnology Information (NCBI), Global Initiative on Sharing All Influenza Data (GISAID), and Virus Pathogen Resource (ViPR), as well as GitHub repositories and other genomic data sources. The visualization integrates sequence data with other data types such as geographic information, serology, or host species in a consistent and concise format accessible to health professionals, epidemiologists, virologists and the public alike 1,2.
Nextstrain displays dashboards developed by both internal maintainers (Nextstrain-maintained pathogen analyses) and community contributors (Scalable Sharing with Nextstrain Groups), all using the same reproducible open-source bioinformatics pipeline 5–7. Nextstrain currently maintains dashboards for the following endemic infections and emerging outbreaks:
RNA Viruses - Respiratory: Avian influenza, HMPV (Human metapneumovirus), Measles, Mumps, RSV (Respiratory syncytial virus), Rubella, SARS-CoV-2, Seasonal CoV (Seasonal coronaviruses), Seasonal influenza
RNA Viruses - Vector-borne (Arboviruses): Dengue, Oropouche, West Nile Virus, Yellow Fever Virus, Zika
RNA Viruses - Hemorrhagic Fever: Ebola, Lassa, Nipah
RNA Viruses - Gastrointestinal: Norovirus
RNA Viruses - Neurotropic: Enterovirus, Rabies
DNA Viruses: Mpox (Monkeypox)
Bacteria: Tuberculosis
Nextstrain offers an open-source toolkit—a collection of core functions and packages—that powers both the insights on their main webpage and enables researchers to analyze their own data. These tools can be accessed and installed for local use at: Installing Nextstrain 8,9.
- Augur: A suite of bioinformatics tools that together provide complete phylogenetic analysis using configurable “recipes” (called builds) specific to different pathogens. Includes composable functions for filtering and aligning sequences, inferring trees, and integrating metadata 10.
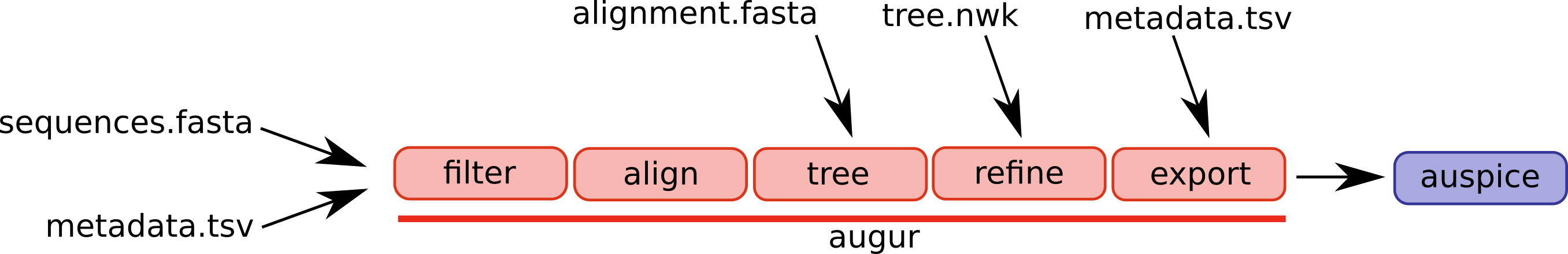
Auspice (Package and Webtool): Dynamic visualization tool for data generated by the Augur pipeline. It powers all pathogen dashboards on Nextstrain, including community-created dashboards 8,11–13.
Nextstrain CLI: The Nextstrain CLI integrates Augur, Auspice, and deployment functionality, providing a unified interface to run pathogen workflows across computing environments (Docker, Conda, AWS Batch) and publish datasets to nextstrain.org or self-hosted pages 14.
Gaining Access
Getting Data
Nextstrain is committed to open science while respecting data privacy and ownership. Researchers can access publicly available sequencing data through dashboards that draw from public repositories, such as GenBank and GISAID where applicable 1. The platform provides derived data, including phylogenetic trees, metadata, and dashboard screenshots that users can download and integrate with their own metadata files 19. Additionally, researchers can view previous data releases or compare different releases. See Viewing previous analyses for detailed guidance 20.
To access publicly available datasets, navigate to any Nextstrain dashboard and scroll to the bottom of the page to locate the download button, or press the “d” key to open the data download popup window. Available download options include:
Phylogenetic Trees: Download newick trees with branch lengths (in terms of genetic divergence or time). Trees include all data and do not reflect current filtering.
Metadata: Per-strain metadata for all samples or current visualization. Includes traits and authorship. An “authors” metadata table is also available.
Diversity (entropy) data: Download data behind the entropy (diversity) panel. The data mirrors what is displayed and includes nucleotide positions and base change counts.
SVG Screenshot: Download a vector image for editing and display. Screenshots are licensed under CC-BY.
Relevant Links
Download the data displayed through nextstrain.org: Find what types of information are available for download. Note that availability may vary between sources 19.
Share Analyses through Nextstrain: Find information on how Nextstrain enables real-time, pre-publication sharing of results 22.
Data: Find documentation on how visualized data is stored and distributed 23.
Viewing previous analyses: Find information about how analysis snapshots are generated, how to navigate historical versions, and how to compare key details available across different time releases of the dashboard data 20.
Nextstrain Documentation: Find information on using and interpreting Nextstrain, their bioinformatics tools and installation guides, and tutorials for genome sequence analysis and phylogenetic workflows 24.
Nextstrain GitHub: Find all public codebases for the project, including data builds for visualization and modeling such as forecasting 25.
Nextstrain Discussions Forum: Find Nextstrain’s community forum where users can ask questions, discuss projects, and share ideas about the platform or research applications 26.
Rapid data analysis and sharing using Nextstrain: The Grubaugh Lab at Yale School of Public Health contributes genomic surveillance data to Nextstrain, a collaborative bioinformatics platform for tracking pathogen evolution and spread. Find their current dashboards visualizing surveillance data for SARS-CoV-2, Dengue, Powassan virus, among others 27.
Nextstrain: real-time tracking of pathogen evolution Hadfiel et al. 2018: Find the article introducing the Nextstrain resource, which highlights the scientific need being addressed, methods, and other important features 1.
Publications
This section presents a selection of PubMed articles that utilize the dataset and are authored by individuals affiliated with the Yale University. These articles are provided to inspire researchers and students to use the data in their own work.
-
Genomic epidemiology of West Nile virus in Europe.
R Tobias Koch, Diana Erazo, Arran J Folly, Nicholas Johnson, Simon Dellicour, Nathan D Grubaugh, Chantal B F Vogels
One health (Amsterdam, Netherlands) 2023 Dec 16 doi: 10.1016/j.onehlt.2023.100664
PMID: 38193029